Gerbil Genome
Original Publication: Zorio DAR, Monsma S, Sanes DH, Golding NL, Rubel EW, Wang Y (2018). De novo sequencing and initial annotation of the Mongolian gerbil ( Meriones unguiculatus ) genome. Genomics pii: S0888-7543(17)30161-1. PDF
Animal, Tissue, and DNA preparation
Mongolian gerbils ( Meriones unguiculatus ), strain 243, were purchased from the Charles River Laboratories (Wilmington, MA). One male animal of 6 weeks of age was used for tissue extraction for this project. Genomic DNA was prepared from leg muscle tissue and quantified using Qubit High Sensitivity reagents.
Library construction, sequencing and assembly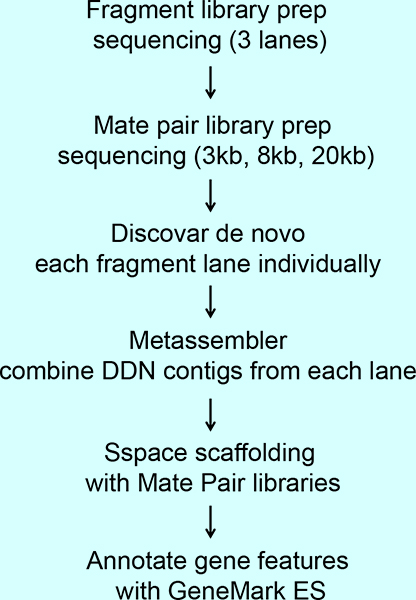
Genomic DNA was ultrasonically sheared for fragment libraries. Fragment libraries were constructed with the NxSeq AmpFree kit (Lucigen), and mate pair libraries with the NxSeq Long Mate Pair kit (Lucigen). Fragment libraries were sequenced on three lanes of HiSeq X (Illumina) with 2x 150 paired end (PE) chemistry at the Hudson Alpha Institute for Biotechnology. Mate pair libraries were sequenced on MiSeq (Illumina) 2x 150 PE with V2 chemistry. Mate pair data was processed with Python scripts Illumina-Chimera-Clean5.py and IlluminaJunctionSplit9.py (available from Lucigen) to remove chimeric mate pairs and to trim the right and left mates by detection of the Junction Code sequence.
Initial assembly was performed with Discovar De Novo using untrimmed fragment data. Each lane of HiSeq X data was assembled individually. The three sets of final Discovar De Novo contigs were then merged into a single assembly by using Metassembler v1.5. Metassembler contigs were then scaffolded sequentially with the 3-, 8- and 20-kb mate pair libraries using Sspace Basic v2.0. Repetitive sequences were identified by using Repeat Modeler v1.08 for de novo repeat discovery. The unmasked assembly was filtered to remove all contigs smaller than 1 kb, and the remainder was deposited with GenBank and submitted for annotation by the NCBI Eukaryotic Genome Annotation Pipeline.
main genome page: https://www.ncbi.nlm.nih.gov/nuccore/NHTI00000000
download sequences: https://www.ncbi.nlm.nih.gov/Traces/wgs/?val=NHTI01#contigs
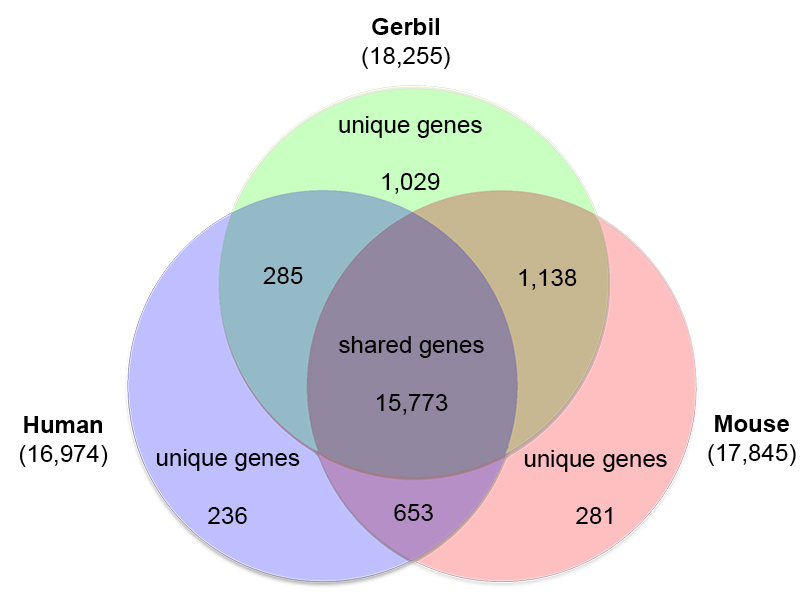 Orthologous gene groups were identified between the Gerbil protein set, the human proteome (UP00005640_9606) and the mouse proteome (UP00000589_10090) from UniProt. The TriFusion v0.5.0 pipeline, which incorporates Usearch for protein-protein comparisons and the OrthoMCL pipeline, was used for proteome comparisons and generation of orthologous protein families. Gene functional annotation clustering was performed with DAVID 6.8 (
https://david.ncifcrf.gov
) using 195 human Uniprot accession numbers for unique orthologs shared between human and gerbil, and the mouse Uniprot accession numbers were used for 760 unique orthologs shared between mouse and gerbil, and 538 unique orthologs shared between mouse and human.
Orthologous gene groups were identified between the Gerbil protein set, the human proteome (UP00005640_9606) and the mouse proteome (UP00000589_10090) from UniProt. The TriFusion v0.5.0 pipeline, which incorporates Usearch for protein-protein comparisons and the OrthoMCL pipeline, was used for proteome comparisons and generation of orthologous protein families. Gene functional annotation clustering was performed with DAVID 6.8 (
https://david.ncifcrf.gov
) using 195 human Uniprot accession numbers for unique orthologs shared between human and gerbil, and the mouse Uniprot accession numbers were used for 760 unique orthologs shared between mouse and gerbil, and 538 unique orthologs shared between mouse and human.


